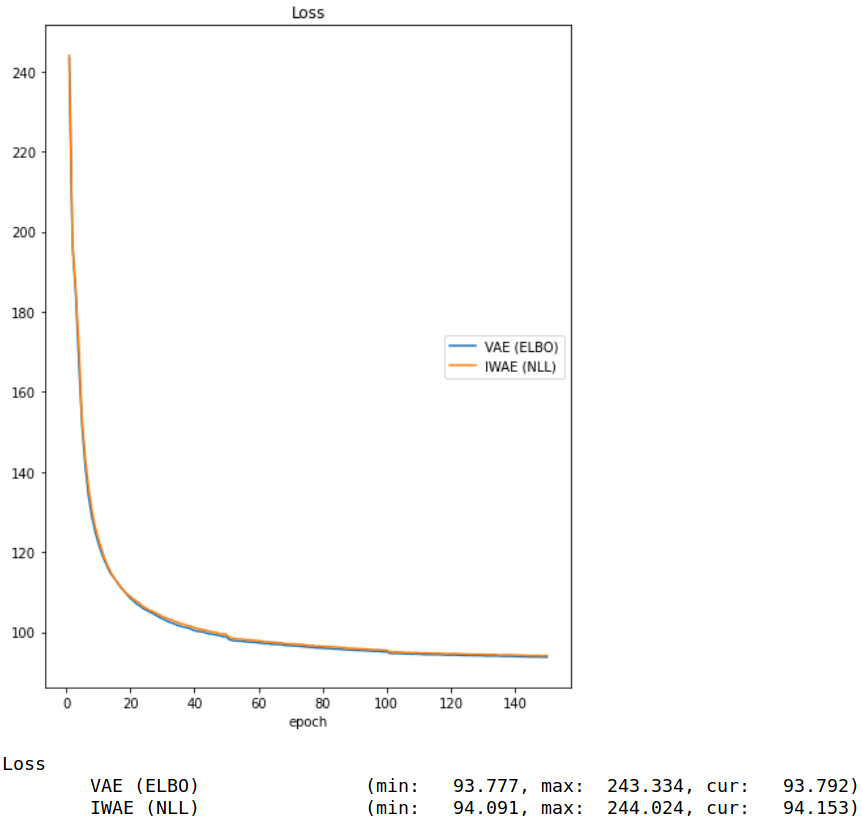
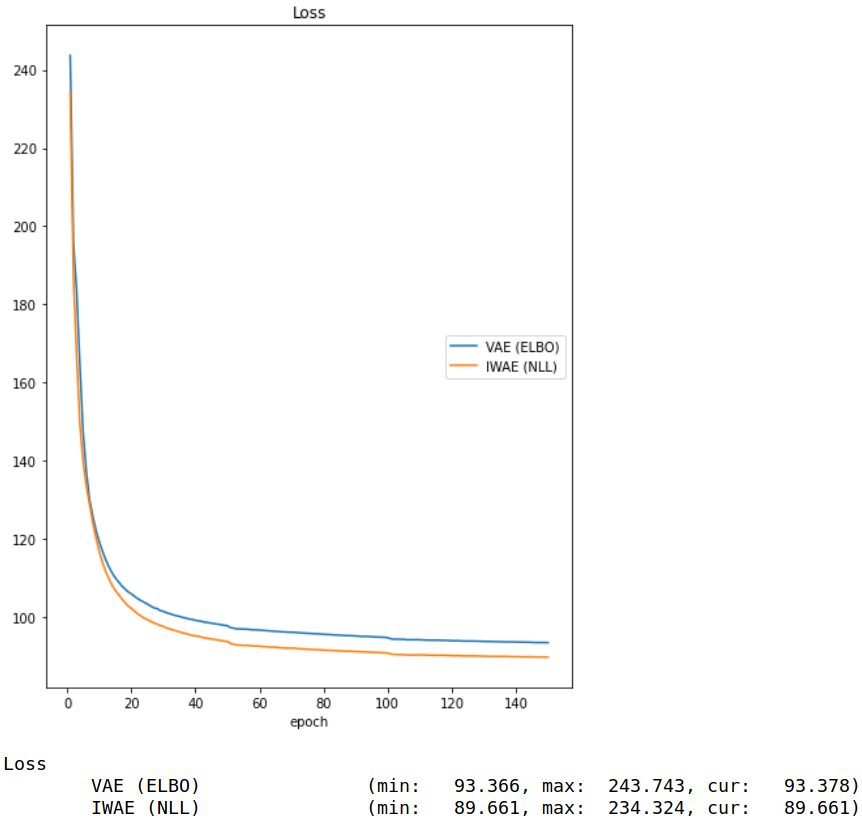
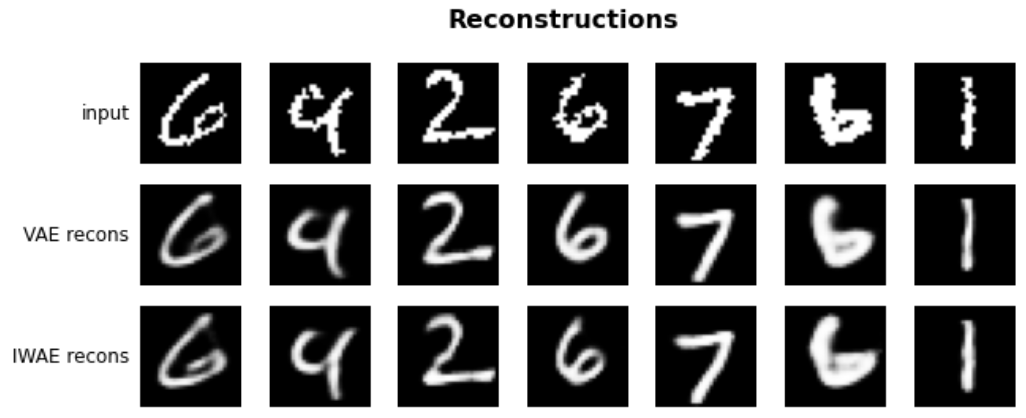
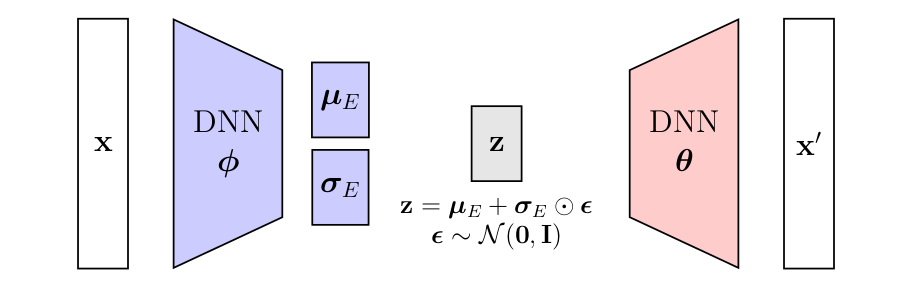IWAE Formulation
Before we introduce the IWAE estimator, remind that the Monte-Carlo estimator of the data likelihood (when the sampling distribution is changed via importance sampling, see Derivation) is given by
\[
p_{\boldsymbol{\theta}} (\textbf{x} ) =
\mathbb{E}_{\textbf{z} \sim q_{\boldsymbol{\phi}} \left( \textbf{z} |
\textbf{x}^{(i)} \right)}
\left[ \frac {p_{\boldsymbol{\theta}} \left(\textbf{x} , \textbf{z}
\right)} {q_{\boldsymbol{\phi}} \left( \textbf{z} | \textbf{x} \right)}
\right] \approx \frac {1}{k} \sum_{l=1}^{k}
\frac {p_{\boldsymbol{\theta}} \left(\textbf{x} , \textbf{z}^{(l)}
\right)} {q_{\boldsymbol{\phi}} \left( \textbf{z}^{(l)} | \textbf{x}
\right)} \quad \text{with} \quad \textbf{z}^{(l)} \sim q_{\boldsymbol{\phi}} \left( \textbf{z} |
\textbf{x}^{(i)} \right)
\]
As a result, the data log-likelihood estimator for one sample \(\textbf{x}^{(i)}\) can be stated as follows
\[
\begin{align}
\log p_{\boldsymbol{\theta}} (\textbf{x}^{(i)} ) &\approx \log \left[ \frac {1}{k} \sum_{l=1}^{k}
\frac {p_{\boldsymbol{\theta}} \left(\textbf{x}^{(i)} , \textbf{z}^{(i, l)}
\right)} {q_{\boldsymbol{\phi}} \left( \textbf{z}^{(i, l)} | \textbf{x}^{(i)}
\right)}\right] = \widetilde{\mathcal{L}}^{\text{IWAE}}_k \left( \boldsymbol{\theta},
\boldsymbol{\phi}; \textbf{x}^{(i)} \right) \\
&\text{with} \quad \textbf{z}^{(i, l)} \sim q_{\boldsymbol{\phi}} \left( \textbf{z} |
\textbf{x}^{(i)} \right)
\end{align}
\]
which leads to an empirical estimate of the IWAE objective. However, Burda et al. (2016) do not use the data log-likelihood in its plain form as the true IWAE objective. Instead they introduce the IWAE objective as follows
\[
\mathcal{L}^{\text{IWAE}}_k \left(\boldsymbol{\theta}, \boldsymbol{\phi};
\textbf{x}^{(i)}\right)
= \mathbb{E}_{\textbf{z}^{(1)}, \dots, \textbf{z}^{(k)} \sim q_{\phi} \left( \textbf{z}|
\textbf{x}^{(i)} \right)}
\left[
\log \frac {1}{k}
\sum_{l=1}^k
\frac {p_{\boldsymbol{\theta}} \left(\textbf{x}^{(i)}, \textbf{z}^{(l)}\right)}
{q_{\phi} \left( \textbf{z}^{(l)} | \textbf{x}^{(i)} \right)}
\right]
\]
For notation purposes, they denote
\[
\text{(unnormalized) importance weights:} \quad
{w}^{(i, l)} = \frac {p_{\boldsymbol{\theta}} \left(\textbf{x}^{(i)}, \textbf{z}^{(l)}\right)}
{q_{\phi} \left( \textbf{z}^{(l)} | \textbf{x}^{(i)} \right)}
\]
By applying Jensen’s Inequality, we can see that in fact the (true) IWAE estimator is merely a lower-bound on the plain data log-likelihood
\[
\mathcal{L}^{\text{IWAE}}_k \left( \boldsymbol{\theta}, \boldsymbol{\phi};
\textbf{x}^{(i)} \right)
= \mathbb{E} \left[ \log \frac {1}{k} \sum_{l=1}^{k} {w}^{(i,
l)}\right] \le \log \mathbb{E} \left[ \frac {1}{k} \sum_{l=1}^{k}
{w}^{(i,l)} \right] = \log p_{\boldsymbol{\theta}} \left( \textbf{x}^{(i)} \right)
\]
They could prove that with increasing \(k\) the lower bound gets strictly tighter and approaches the true data log-likelihood in the limit of \(k \rightarrow
\infty\). Note that since the empirical IWAE estimator \(\widetilde{\mathcal{L}}_k^{\text{IWAE}}\) can be understood as a Monte-Carlo estimator on the true data log-likelihood, in the empirical case this property can simply be deduced from the properties of Monte-Carlo integration.
A very well explanation is given by Domke and Sheldon (2018). Starting from the property
\[
p(\textbf{x}) = \mathbb{E} \Big[ w \Big] = \mathbb{E}_{\textbf{z} \sim q_{\boldsymbol{\phi}} \left( \textbf{z} |
\textbf{x}^{(i)} \right)}
\left[ \frac {p_{\boldsymbol{\theta}} \left(\textbf{x} , \textbf{z}
\right)} {q_{\boldsymbol{\phi}} \left( \textbf{z} | \textbf{x} \right)}
\right]
\]
We derived the ELBO using Jensen’s inequality
\[
\log p(\textbf{x}) \ge \mathbb{E} \Big[ \log w \Big] = \text{ELBO} \Big[ q ||
p \Big]
\]
Suppose that we could make \(w\) more concentrated about its mean \(p(\textbf{x})\). Clearly, this would yield a tighter lower bound when applying Jensen’s Inequality.
(rhetorical break)
Can we make \(w\) more concentrated about its mean? YES, WE CAN.
For example using the sample average \(w_k = \frac {1}{k}
\sum_{i=1}^k w^{(i)}\). This leads directly to the true IWAE objective
\[
\log p(\textbf{x}) \ge \mathbb{E} \Big[ \log w_k \Big] = \mathbb{E} \left[
\log \frac {1}{k} \sum_{i=1}^{k} w^{(i)} \right] = \mathcal{L}^{\text{IWAE}}_k
\]
Here it gets interesting. A closer analysis on the IWAE bound by Nowozin (2018) revealed the following property
\[
\begin{align}
&\quad \mathcal{L}_k^{\text{IWAE}} = \log p(\textbf{x}) - \frac {1}{k} \frac
{\mu_2}{2\mu^2} + \frac {1}{k^2} \left( \frac {\mu_3}{3\mu^3} - \frac
{3\mu_2^2}{4\mu^4} \right) + \mathcal{O}(k^{-3})\\
&\text{with} \quad
\mu = \mathbb{E}_{\textbf{z} \sim q_{\boldsymbol{\phi}}} \left[ \frac
{p_{\boldsymbol{\theta}}\left( \textbf{x}, \textbf{z}
\right)}{q_{\boldsymbol{\phi}} \left( \textbf{z} | \textbf{x} \right)} \right]
\quad
\mu_i = \mathbb{E}_{\textbf{z} \sim q_{\boldsymbol{\phi}}} \left[
\left( \frac
{p_{\boldsymbol{\theta}}\left( \textbf{x}, \textbf{z}
\right)}{q_{\boldsymbol{\phi}} \left( \textbf{z} | \textbf{x} \right)}
- \mathbb{E}_{\textbf{z} \sim q_{\boldsymbol{\phi}}} \left[ \frac
{p_{\boldsymbol{\theta}}\left( \textbf{x}, \textbf{z}
\right)}{q_{\boldsymbol{\phi}} \left( \textbf{z} | \textbf{x} \right)} \right]
\right)^2 \right]
\end{align}
\]
Thus, the true objective is a biased - in the order of \(\mathcal{O}\left(k^{-1}\right)\) - and consistent estimator of the marginal log likelihood \(\log p(\textbf{x})\). The empirical estimator of the true IWAE objective is basically a special Monte-Carlo estimator (only one sample per \(k\)) on the true IWAE objective. It is more or less luck that we can formulate the same empirical objective and interpret it differently as the Monte-Carlo estimator (with \(k\) samples) on the data log-likelihood.
Let us take a closer look on how to compute gradients (fast) for the empirical estimate of the IWAE objective:
\[
\begin{align}
\nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}}
\widetilde{\mathcal{L}}_k^{\text{IWAE}} \left( \boldsymbol{\theta}, \boldsymbol{\phi};
\textbf{x}^{(i)} \right) &= \nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}}
\log \frac {1}{k} \sum_{l=1}^k w^{(i,l)} \left( \textbf{x}^{(i)},
\textbf{z}^{(i, l)}_{\boldsymbol{\phi}}, \boldsymbol{\theta} \right) \quad
\text{with} \quad
\textbf{z}^{(i, l)} \sim q_{\boldsymbol{\phi}} \left(\textbf{z} |
\textbf{x}^{(i)} \right)\\
&\stackrel{\text{(*)}}{=}
\sum_{l=1}^{k} \frac {w^{(i, l)}}{\sum_{m=1}^{k} w^{(i,
m)}} \nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}} \log w^{(i,l)} =
\sum_{l=1}^{k} \widetilde{w}^{(i, l)} \nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}} \log w^{(i,l)},
\end{align}
\]
where we introduced the following notation
\[
\text{(normalized) importance weights:} \quad
\widetilde{w}^{(i, l)} = \frac {w^{(i,l)}}{\sum_{m=1}^k w^{(i, m)}}
\]
\[
\begin{align}
\frac {\partial \left[ \log \frac {1}{k} \sum_i^{k} w_i \left( \boldsymbol{\theta}
\right) \right]}{\partial \boldsymbol{\theta}} &\stackrel{\text{chain rule}}{=} \frac {\partial
\log a}{\partial a} \sum_{i}^{k} \frac {\partial a}{\partial w_i} \frac
{\partial w_i}{\partial \boldsymbol{\theta}} \quad \text{with}
\quad a = \frac {1}{k} \sum_{i}^k w_i (\boldsymbol{\theta})\\
&= \frac {k}{\sum_l^k w_l} \sum_{i}^{k}\frac {1}{k} \frac {\partial
w_i (\boldsymbol{\theta})}{\partial \boldsymbol{\theta}} = \frac {1}{\sum_l^k
w_l} \sum_{i}^{k} \frac {\partial
w_i (\boldsymbol{\theta})}{\partial \boldsymbol{\theta}}
\end{align}
\]
Lastly, we use the following identity
\[
\frac {\partial w_i (\boldsymbol{\theta})}{\partial \boldsymbol{\theta}} = w_i
(\boldsymbol{\theta}) \cdot
\frac {\partial \log w_i (\boldsymbol{\theta})}{\partial \boldsymbol{\theta}}
\stackrel{\text{chain rule}}{=} w_i (\boldsymbol{\theta}) \cdot \frac {1}{w_i
(\boldsymbol{\theta})} \cdot
\frac {\partial w_i (\boldsymbol{\theta})}{\partial \boldsymbol{\theta}} =
\frac {\partial w_i (\boldsymbol{\theta})}{\partial \boldsymbol{\theta}}
\]
Similar to VAEs, this formulation poses a problem for backpropagation due to the sampling operation. We use the same reparametrization trick to circumvent this problem and obtain a low variance update rule:
\[
\begin{align}
\nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}}
\widetilde{\mathcal{L}}_k^{\text{IWAE}} &=
\sum_{l=1}^{k} \widetilde{w}^{(i, l)} \nabla_{\boldsymbol{\phi},
\boldsymbol{\theta}} \log w^{(i,l)} \left( \textbf{x}^{(i)},
\textbf{z}_{\boldsymbol{\phi}}^{(i,l)}, \boldsymbol{\theta} \right)
\quad \text{with} \quad
\textbf{z}^{(i,l)} \sim q_{\boldsymbol{\phi}} \left(\textbf{z} | \textbf{x}^{(i)} \right)\\
&= \sum_{l=1}^k \widetilde{w}^{(i,l)} \nabla_{\boldsymbol{\phi},
\boldsymbol{\theta}} \log w^{(i,l)} \left(\textbf{x}^{(i)},
g_{\boldsymbol{\phi}} \left( \textbf{x}^{(i)},
\boldsymbol{\epsilon}^{(l)}\right), \textbf{x}^{(i)} \right), \quad \quad
\boldsymbol{\epsilon} \sim p(\boldsymbol{\epsilon})
\end{align}
\]
To make things clearer for the implementation, let us unpack the log
\[
\log w^{(i,l)} = \log \frac {p_{\boldsymbol{\theta}} \left(\textbf{x}^{(i)}, \textbf{z}^{(l)}\right)}
{q_{\boldsymbol{\phi}} \left( \textbf{z}^{(l)} | \textbf{x}^{(i)} \right)} = \underbrace{\log
p_{\boldsymbol{\theta}} \left (\textbf{x}^{(i)} | \textbf{z}^{(l)}
\right)}_{\text{NLL}} + \log p_{\boldsymbol{\theta}} \left( \textbf{z}^{(l)}
\right) - \log q_{\boldsymbol{\phi}} \left( \textbf{z}^{(l)} | \textbf{x}^{(i)} \right)
\]
Before, we are going to implement this formulation, let us look whether we can separate out the KL divergence for the true IWAE objective of Burda et al. (2016). Therefore, we state the update for the true objective:
\[
\begin{align}
\nabla_{\boldsymbol{\phi},
\boldsymbol{\theta}}
\mathcal{L}_k^{\text{IWAE}} &=
\nabla_{\boldsymbol{\phi},
\boldsymbol{\theta}}
\mathbb{E}_{\textbf{z}^{(1)}, \dots, \textbf{z}^{(l)}} \left[ \log \frac {1}{k}
\sum_{l=1}^{k} w^{(l)} \left( \textbf{x},
\textbf{z}^{(l)}_{\boldsymbol{\phi}}, \boldsymbol{\theta} \right) \right]\\
&=
\mathbb{E}_{\textbf{z}^{(1)}, \dots, \textbf{z}^{(l)}} \left[
\sum_{l=1}^{k} \widetilde{w}_i
\nabla_{\boldsymbol{\phi},
\boldsymbol{\theta}}
\log w^{(l)} \left( \textbf{x}, \textbf{z}_{\boldsymbol{\phi}}^{(l)}, \boldsymbol{\theta} \right) \right]\\
&=\sum_{l=1}^{k} \widetilde{w}_i \mathbb{E}_{\textbf{z}^{(l)}} \left[
\nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}} \log w^{(l)} \left( \textbf{x},
\textbf{z}_{\boldsymbol{\phi}}^{(l)}, \boldsymbol{\theta} \right)
\right]\\
&\neq \sum_{l=1}^{k} \widetilde{w}_i
\nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}}
\mathbb{E}_{\textbf{z}^{(l)}} \left[
\log w^{(l)} \left( \textbf{x},
\textbf{z}_{\boldsymbol{\phi}}^{(l)}, \boldsymbol{\theta} \right)
\right]
\end{align}
\]
Unfortunately, we cannot simply move the gradient outside the expectation. If we could, we could simply rearrange the terms inside the expectation as in the standard VAE case.
Let us look, what would happen, if we were to describe the true IWAE estimator as the data log-likelihood \(\log p \left( \textbf{x} \right)\) in which the sampling distribution is exchanged via importance sampling:
\[
\begin{align}
\nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}} \log p \left( \textbf{x}^{(i)} \right) &=
\nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}} \log \mathbb{E}_{\textbf{z} \sim
q_{\boldsymbol{\phi}} \left( \textbf{z} | \textbf{x}^{(i)}\right)} \left[ w
(\textbf{x}^{(i)}, \textbf{z}, \boldsymbol{\theta})\right]\\
&\neq
\nabla_{\boldsymbol{\phi}, \boldsymbol{\theta}} \mathbb{E}_{\textbf{z} \sim
q_{\boldsymbol{\phi}} \left( \textbf{z} | \textbf{x}^{(i)}\right)} \left[ \log w
(\textbf{x}^{(i)}, \textbf{z}, \boldsymbol{\theta})\right]
\end{align}
\]
Here, we also cannot separate the KL divergence out, since we cannot simply move the log inside the expectation.