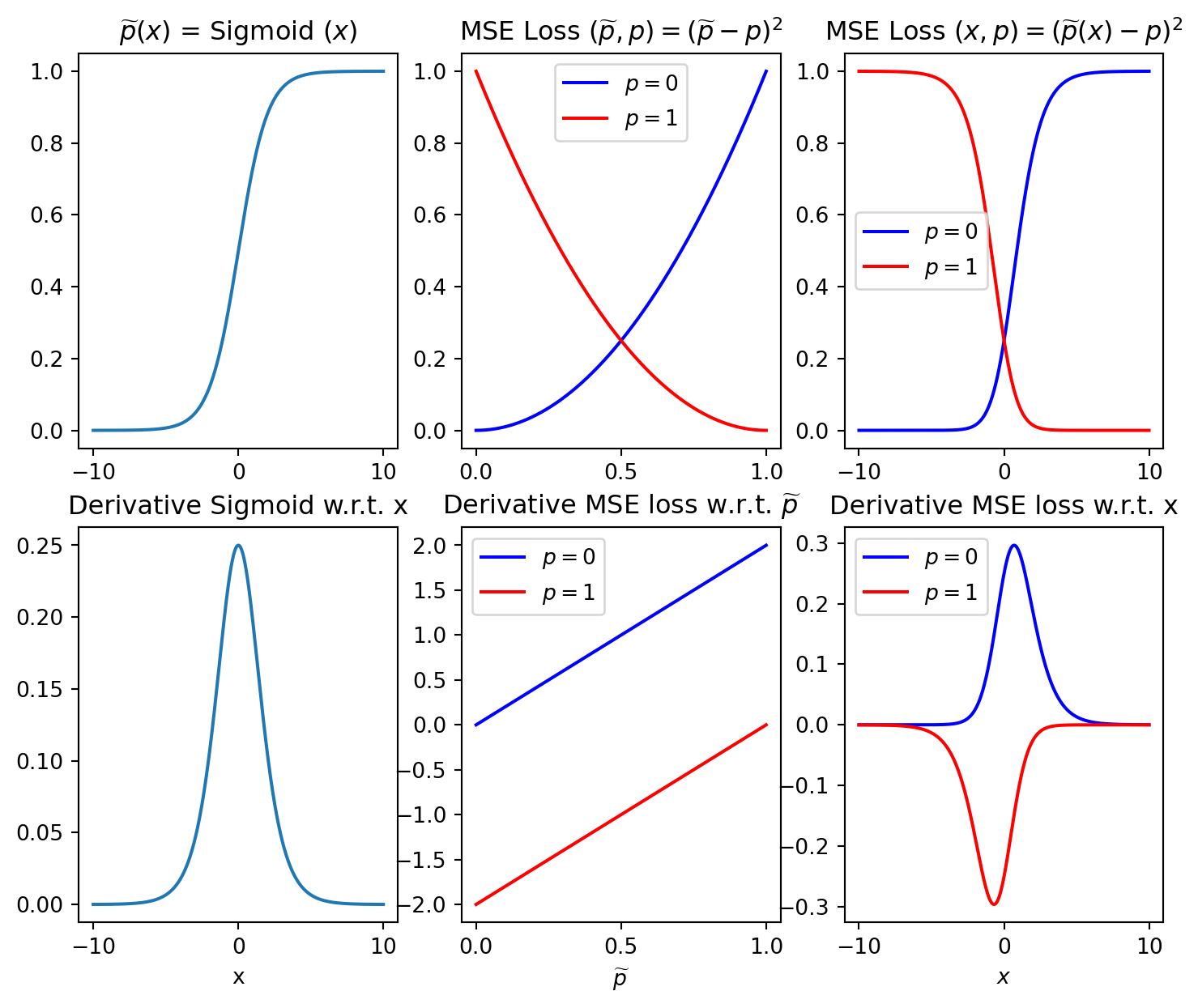
Let’s denote the output of the network by \(\widetilde{\textbf{p}} \in [0, 1]^{N}\) (for estimated probabilities) and the input to the sigmoid \(\textbf{x}\in \mathbb{R}^{N}\) (i.e., layer output before sigmoid is applied)
\[
\textbf{p} = \text{Sigmoid} (\textbf{x}) = \frac {1}{1 + \exp \left({-\textbf{x}}\right)}
\]
Now suppose that \(\textbf{p}\in [0, 1]^{N}\) denotes the true probabilities, then applying the MSE loss gives
\[
\text{MSE loss} \left(\widetilde{\textbf{p}}, \textbf{p}\right)
=J\left(\widetilde{\textbf{p}}, \textbf{p}\right) = \sum_{i=1}^N
\left(\widetilde{p}_i - p_i \right)^2,
\]
The gradient w.r.t. network parameters will be proportional to the gradient w.r.t. \(\textbf{x}\) (backpropagation rule) which is as follows
\[
\frac {\partial J \left(\widetilde{\textbf{p}}, \textbf{p}\right)}{\partial
x_i} = \sum_{j=1}^N \frac {\partial J}{\partial p_j} \cdot \frac {\partial p_j}{\partial x_j} = 2
\left(\widetilde{p}_i - p_i \right) \cdot \frac {\partial p_i}{\partial x_i} = 2
\left(\widetilde{p}_i - p_i \right) \cdot p_i (1 - p_i)
\]
Here we can directly see that even if the absolute error approaches 1, i.e., \(\left(\widetilde{p}_i - p_i \right)\rightarrow 1\), the gradient vanishes for \(p_i\rightarrow 1\) and \(p_i\rightarrow 0\).
Let’s derive the gradient of the sigmoid using substitution and the chain rule
\[
\begin{align}
\frac {\partial p_i} {\partial x_i} &= \frac {\partial u}{\partial t} \cdot \frac
{\partial t}{\partial x} \quad \text{with} \quad u(t) = t^{-1}, \quad t(x_i) = 1 +
\exp{(-x_i)}\\
&= -t^{-2} \cdot \left(-\exp{x_i}\right) = \frac {exp\left(-x_i\right)}{\left(1 +
\exp\left(-x_i\right)\right)^2}\\
&= \underbrace{\frac {1}{1 + \exp \left( -x \right)}}_{p_i} \underbrace{\frac {\exp
(-x_i)}{1+exp(-x_i)}}_{1-p_i} = p_i \left(1 - p_i \right)
\end{align}
\]